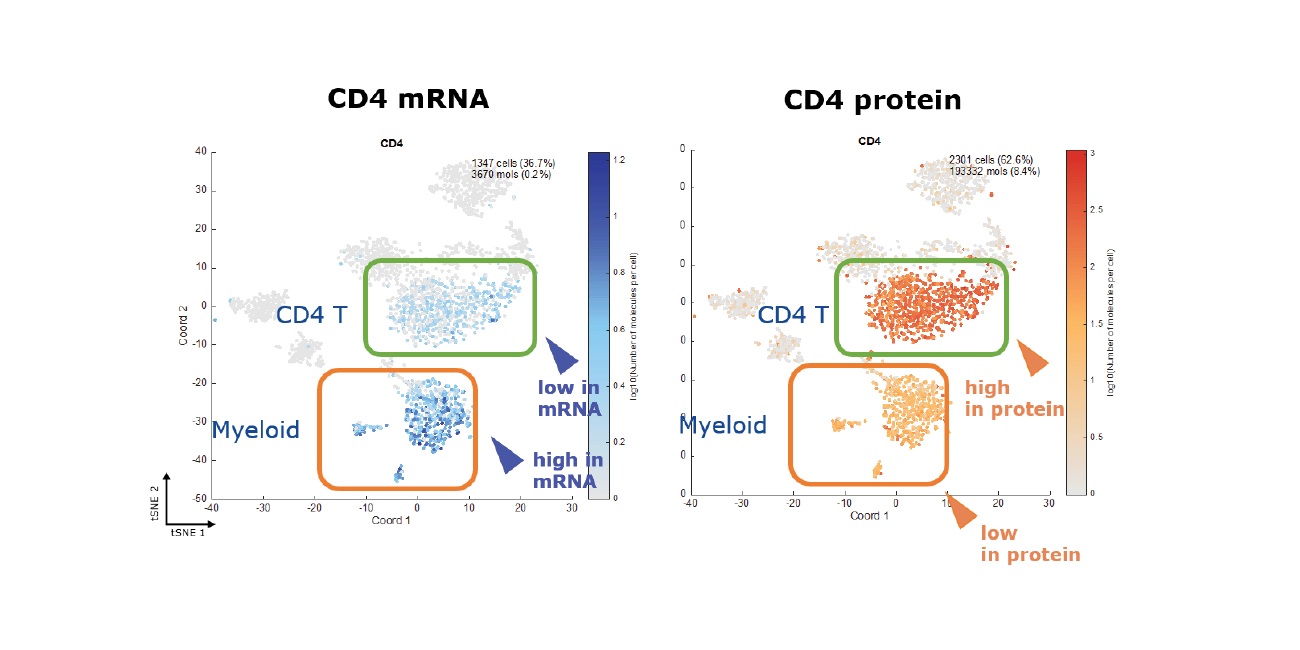
Ever wonder what would come to light if you could see mRNA and protein expression at the same time?
What would you learn that single-cell RNA-Seq or flow cytometry alone is not showing you?
Techniques for differential gene expression analysis, such as qPCR, arrays and mRNA sequencing, have been important tools for growing our knowledge of biology over the past 20 or so years. The ability to study the same biology through sequencing-based methods such as RNA-Seq, when combined with new, high-throughput single-cell capture technologies has greatly increased the resolution such that we can now more easily analyze mRNA expression in individual cells. This has been a very important advancement towards achieving a greater understanding of complex tissues and disease states. However, prior studies have shown that the correlation between mRNA and protein expression levels is not always predictable, sometimes leaving important gaps that prevent our complete understanding of cellular state or change. The inability to investigate how mRNA and protein expression levels differ in the same cell or between cells within the same sample could mean that we are missing information that is important to understanding the biology of a given system.
So what now?
The ability to combine high-parameter protein expression and single cell RNAseq data would significantly improve our understanding of individual cells and cell populations. Traditional single-cell protein assays, like flow cytometry and microscopy, are limited in the number of parameters that can be achieved within one experiment, and current techniques for combining mRNA and protein expression data in single cells tend to be lower in throughput and more expensive per cell. Researchers have long hoped for a simple and convenient way to achieve this analysis at a cell and target scale that is meaningful and BD has answered the call for a solution.